Postdoctoral position studying critical zone processes in drylands
October 22, 2020 – 12:55 pmI’ve been involved in launching a big new research project at UTEP focused on critical zone processes in drylands led by my colleague in geology Dr. Lixin Jin. We’re currently recruiting a postdoc to work especially on the eddy flux tower components of the project. I’ll be the main point of contact at UTEP for this part of the project and the postdoc will work with me, but the position is stationed in Boise, Idaho and will also be heavily collaborative with Gerald Flerchinger at USDA, who runs a set of eddy towers associated with the Reynolds Creek site, and Jen Pierce at Boise State. As the ad says, please write an email to all of us if you are interested. It’s a cool project with tons of collaborators and opportunities to get involved with the critical zone research community.
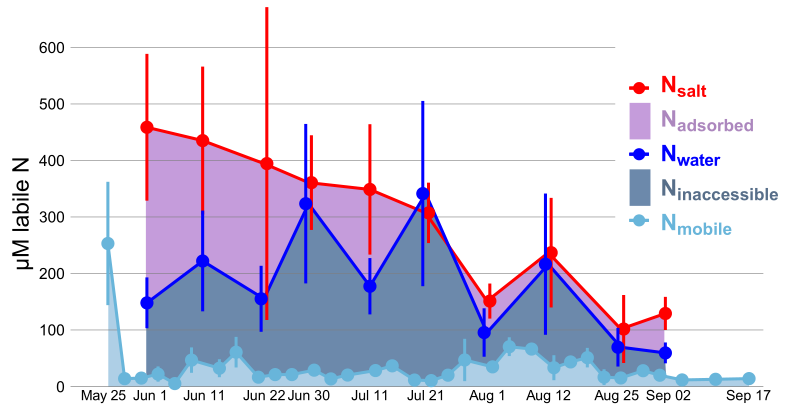
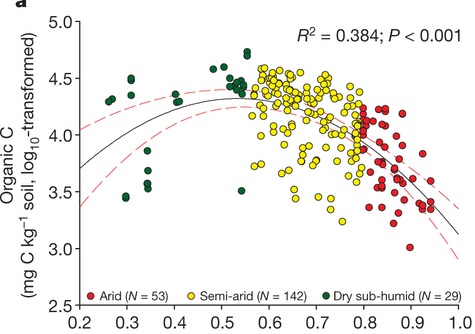
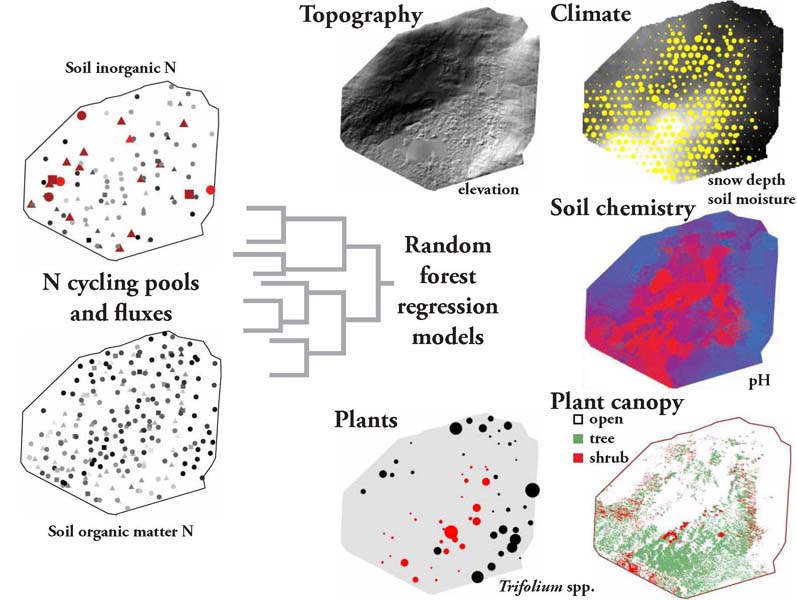
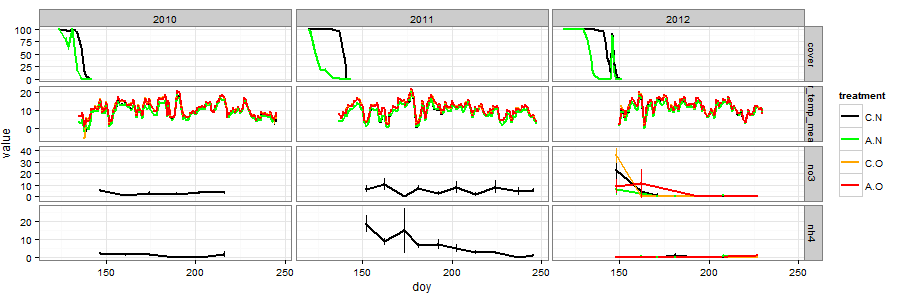
The University of Texas at El Paso (UTEP), in association with Boise State University, and the U.S.D.A. Agricultural Research Service (ARS), is recruiting a postdoctoral researcher to work on a recently funded National Science Foundation ‘Critical Zone Thematic Cluster’ grant to study carbon fluxes, ecohydrology, and nutrient availability in the carbonate-dominated soils of dryland ecosystems. The project has sites in Texas, New Mexico, and Idaho. While funded through UTEP, this position is located in Boise, Idaho and focused on the scientific field operations at the Idaho sites, including the Reynolds Creek Experimental Watershed and the Northwest Irrigation and Soils Research site in Kimberly, Idaho. We are specifically looking for a candidate who has experience working with eddy covariance towers and the data that they generate. Other useful areas of interest include soil science, soil biogeochemistry, knowledge of dryland ecosystem structure and function, and ecosystem-atmosphere gas exchange techniques. We seek a colleague who is interested in collaborative engagement with scientists and students and also in education and outreach activities. Up to four years of funding is available. We will begin to review applications on Nov 15th but the position is open until filled. The target start date for the position is January 4, 2021. For more information, please email Anthony Darrouzet-Nardi (ajdarrouzetnardi@utep.edu), and copy Lixin Jin (ljin2@utep.edu), Jennifer Pierce (jenpierce@boisestate.edu), and Gerald Flerchinger (gerald.flerchinger@usda.gov). To submit an application, please visit this link and send one pdf file with a cover letter, CV, and contact information of at least three referees.
That link at the bottom is to the official listing on the UTEP HR site. If you have any issues, try clicking it a couple times. The system can be a bit clunky.
Comments Off on Postdoctoral position studying critical zone processes in drylands